Research Progress in the Identification of RNA N6-methyladenosine Modification Sites
Editor: | Apr 08,2024
N6-methyladenosine (m6A), one of the most prevalent RNA modifications, serves as a pivotal epigenetic signal, profoundly influencing splicing regulation, mRNA stability, translation efficiency, and epigenetic modulation. Despite the development of various computational models aimed at accurately identifying potential m6A modification sites, many lack the capacity to elucidate the underlying recognition process through consensus knowledge.
To overcome this problem, the study group from the Xinjiang Technical Institute of Physics and Chemistry proposes a deep learning model, namely M6A-DCR, by discovering consensus regions for interpretable identification of m6A modification sites. In particular, M6A-DCR first constructs an instance graph for each RNA sequence by integrating specific positions and types of nucleotides. The discovery of consensus regions is then formulated as a graph clustering problem in light of aggregating all instance graphs. After that, M6A-DCR adopts a motif-aware graph reconstruction optimization process to learn high-quality embeddings of input RNA sequences, thus achieving the identification of m6A modification sites in an end-to-end manner. Experimental results demonstrate the superior performance of M6A-DCR by comparing it with several state-of-the-art identification models. The consideration of consensus regions empowers our model to make interpretable predictions at the motif level. The analysis of cross validation through different species and tissues further verifies the consistency between the identification results of M6A-DCR and the evolutionary relationships among species, and the potential associations of related effector proteins with the found consensus regions (Figure 1).
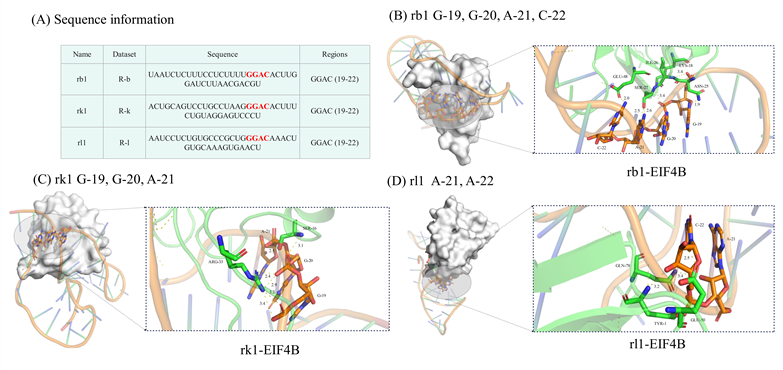
Figure 1. The docking details of the effector protein and the consensus regions This study provides valuable insights for the computational identification of RNA m6A modification sites. The research findings, titled "Discovering consensus regions for interpretable identification of RNA N6-methyladenosine modification sites via graph contrastive clustering" have been published in the IEEE Journal of Biomedical and Health Informatics (Volume: 28, Issue: 4, Pages: 2362-2372, April 2024), and have been featured as the cover article (Figure 2). Xinjiang Technical Institute of Physics and Chemistry was the sole institution involved, with Hu Lun as the corresponding author, and Li Guodong, a Ph.D. graduate from the class of 2022, serving as the primary author. The IEEE Journal of Biomedical and Health Informatics is a premier journal in the Chinese Academy of Sciences Region 1, boasting a current impact factor of 7.7. The research received support from the National Natural Science Foundation of China, the Natural Science Foundation of Xinjiang Uygur Autonomous Region, the Pioneer Hundred Talents Program of Chinese Academy of Sciences and the CAS Light of the West Multidisciplinary Team project. Figure 2. The article cover
附件下载:
 (86) 991-3838931
(86) 991-3838931 lhskj@ms.xjb.ac.cn
lhskj@ms.xjb.ac.cn (86)991-3838957
(86)991-3838957 40-1 Beijing Road
Urumqi, XinjiangChina
40-1 Beijing Road
Urumqi, XinjiangChina